Highlighted Publications
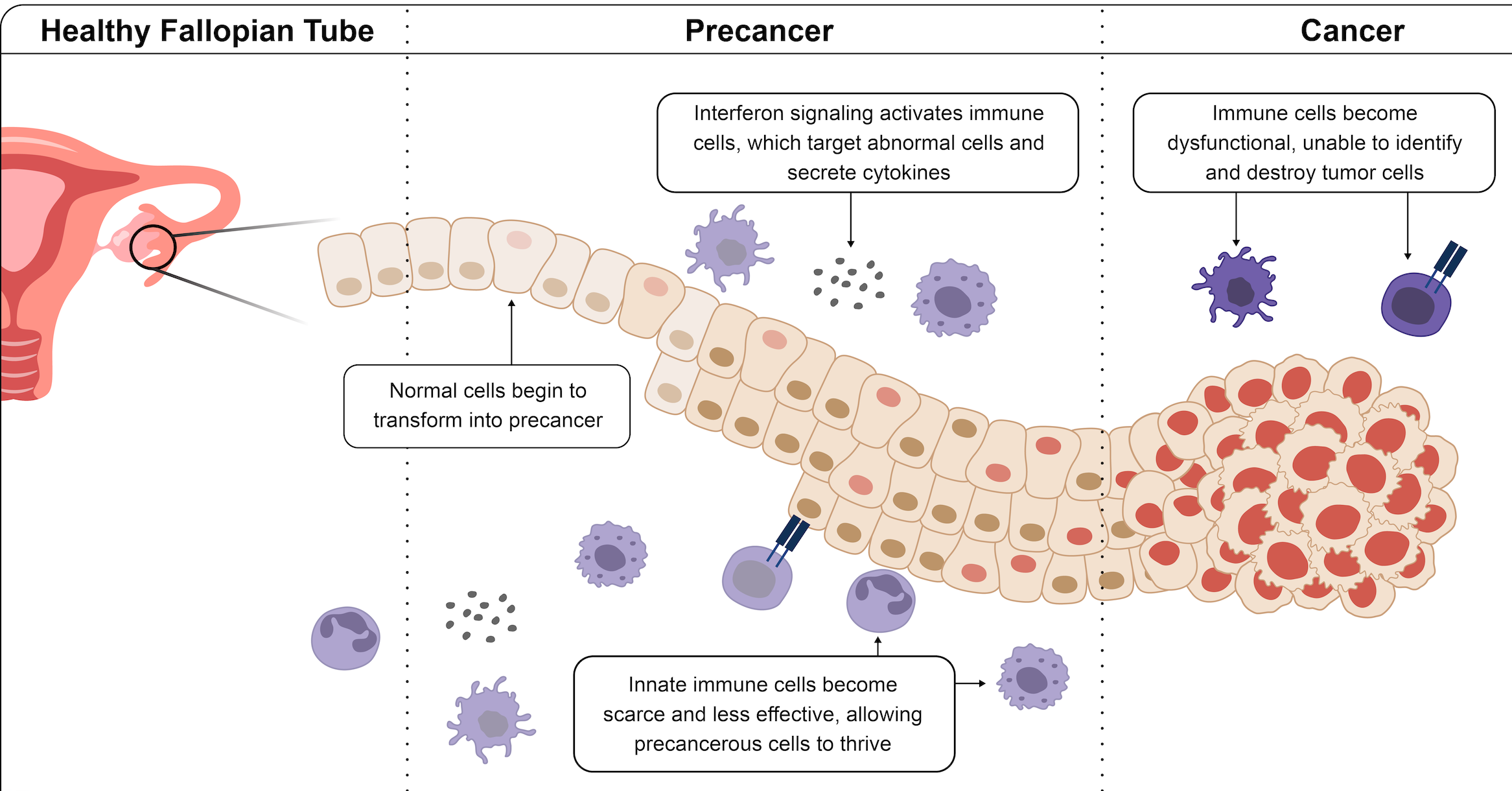
Multimodal Spatial Profiling Reveals Immune Suppression and Microenvironment Remodeling in Fallopian Tube Precursors to High-Grade Serous Ovarian Carcinoma
Kader T, Lin JR, Hug CB, Coy S, Chen YA, de Bruijn I, Shih N, Jung E, Pelletier RJ, Lopez Leon M, Mingo G, Omran DK, Lee JS, Yapp C, Satravada BA, Kundra R, Xu Y, Chan S, Tefft JB, Muhlich JL, Kim SH, Gysler SM, Agudo J, Heath JR, Schultz N, Drescher CW, Sorger PK, Drapkin R, Santagata S.
Cancer Discovery. 2025 May 5. PMID: 39704522.

Epigenetic and Oncogenic Inhibitors Cooperatively Drive Differentiation and Kill KRAS-Mutant Colorectal Cancers
Loi P, Schade AE, Rodriguez CL, Krishnan A, Perurena N, Nguyen VTM, Xu Y, Watanabe M, Davis RA, Gardner A, Pilla NF, Mattioli K, Popow O, Gunduz N, Lannagan TRM, Fitzgerald S, Sicinska ET, Lin JR, Tan W, Brais LK, Haigis KM, Giannakis M, Ng K, Santagata S, Helin K, Sansom OJ, Cichowski K.
Cancer Discov. 2024 Dec 2. PMCID: PMC11609823
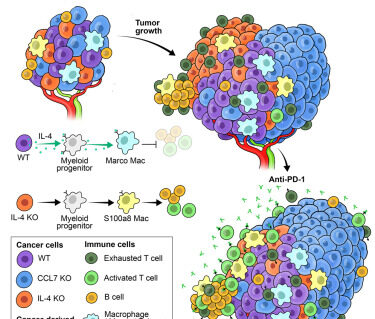
Ovarian cancer-derived IL-4 promotes immunotherapy resistance
Mollaoglu G, Tepper A, Falcomatà C, Potak HT, Pia L, Amabile A, Mateus-Tique J, Rabinovich N, Park MD, LaMarche NM, Brody R, Browning L, Lin JR, Zamarin D, Sorger PK, Santagata S, Merad M, Baccarini A, Brown BD.
Cell. 2024 Oct 25. PMID: 39481380
AKT and EZH2 inhibitors kill TNBCs by hijacking mechanisms of involution
Schade AE, Perurena N, Yang Y, Rodriguez CL, Krishnan A, Gardner A, Loi P, Xu Y, Nguyen VTM, Mastellone GM, Pilla NF, Watanabe M, Ota K, Davis RA, Mattioli K, Xiang D, Zoeller JJ, Lin JR, Morganti S, Garrido-Castro AC, Tolaney SM, Li Z, Barbie DA, Sorger PK, Helin K, Santagata S, Knott SRV, Cichowski K.
Nature. 2024 Oct 9; PMID: 39385030

Nociceptor-immune interactomes reveal insult-specific immune signatures of pain
Jain A, Gyori BM, Hakim S, Jain A, Sun L, Petrova V, Bhuiyan SA, Zhen S, Wang Q, Kawaguchi R, Bunga S, Taub DG, Ruiz-Cantero MC, Tong-Li C, Andrews N, Kotoda M, Renthal W, Sorger PK, Woolf CJ.
Nat Immunol. 2024 May 28. PMCID: PMC11224023

Phase II Study of Eribulin plus Pembrolizumab in Metastatic Soft Tissue Sarcomas: Clinical Outcomes and Biological Correlates
Haddox CL, Nathenson MJ, Mazzola E, Lin JR, Baginska J, Nau A, Weirather JL, Choy E, Marino-Enriquez A, Morgan JA, Cote GM, Merriam P, Wagner AJ, Sorger PK, Santagata S, George S.
Clin Cancer Res. 2024 Jan 18. PMCID: PMC10982640

Targeting TBK1 to overcome resistance to cancer immunotherapy
Sun Y, Revach OY, Anderson S, Kessler EA, Wolfe CH, Jenney A, Mills CE, Robitschek EJ, Davis TGR, Kim S, Fu A, Ma X, Gwee J, Tiwari P, Du PP, Sindurakar P, Tian J, Mehta A, Schneider AM, Yizhak K, Sade-Feldman M, LaSalle T, Sharova T, Xie H, Liu S, Michaud WA, Saad-Beretta R, Yates KB, Iracheta-Vellve A, Spetz JKE, Qin X, Sarosiek KA, Zhang G, Kim JW, Su MY, Cicerchia AM, Rasmussen MQ, Klempner SJ, Juric D, Pai SI, Miller DM, Giobbie-Hurder A, Chen JH, Pelka K, Frederick DT, Stinson S, Ivanova E, Aref AR, Paweletz CP, Barbie DA, Sen DR, Fisher DE, Corcoran RB, Hacohen N, Sorger PK, Flaherty KT, Boland GM, Manguso RT, Jenkins RW.
Nature. 2023 Mar. PMCID: PMC10171827
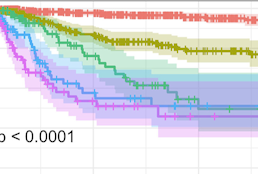
Development and validation of time-to-event models to predict metastatic recurrence of localized cutaneous melanoma
Wan G, Leung BW, DeSimone MS, Nguyen N, Rajeh A, Collier MR, Rashdan H, Roster K, Zhou X, Moseley CB, Nirmal AJ, Pelletier RJ, Maliga Z, Marko-Varga G, Németh IB, Tsao H, Asgari MM, Gusev A, Stagner AM, Lian CG, Hurlbert MS, Liu F, Yu KH, Sorger PK, Semenov YR.
J Am Acad Dermatol. 2023 Oct 3;S0190-9622(23)02881–5. PMID: 37797836
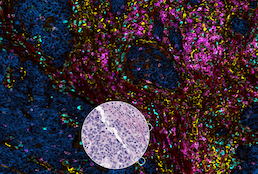
High-plex immunofluorescence imaging and traditional histology of the same tissue section for discovering image-based biomarkers
Lin JR, Chen YA, Campton D, Cooper J, Coy S, Yapp C, Tefft JB, McCarty E, Ligon KL, Rodig SJ, Reese S, George T, Santagata S, Sorger PK
Nature Cancer. 2023 Jul. PMID: 37349501
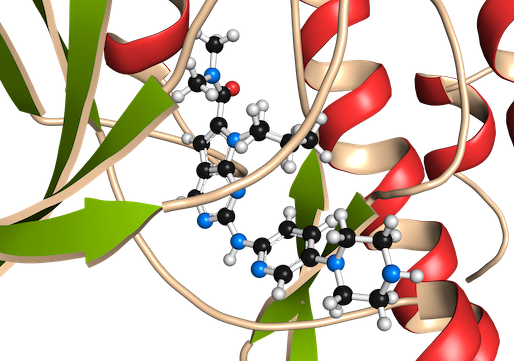
OpenFold: retraining AlphaFold2 yields new insights into its learning mechanisms and capacity for generalization
Ahdritz G, Bouatta N, Floristean C, Kadyan S, Xia Q, Gerecke W, O’Donnell TJ, Berenberg D, Fisk I, Zanichelli N, Zhang B, Nowaczynski A, Wang B, Stepniewska-Dziubinska MM, Zhang S, Ojewole A, Guney ME, Biderman S, Watkins AM, Ra S, Lorenzo PR, Nivon L, Weitzner B, Ban YEA, Chen S, Zhang M, Li C, Song SL, He Y, Sorger PK, Mostaque E, Zhang Z, Bonneau R, AlQuraishi M.
Nat Methods. 2024 May 14. PMID: 38744917
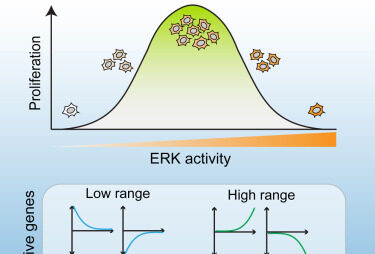
Multi-range ERK responses shape the proliferative trajectory of single cells following oncogene induction.
Chen JY, Hug C, Reyes J, Tian C, Gerosa L, Fröhlich F, Ponsioen B, Snippert HJG, Spencer SL, Jambhekar A, Sorger PK, Lahav G.
Cell Rep. 2023 Mar 28;42(3):112252. PMCID: PMC10153468

Multiplexed 3D atlas of state transitions and immune interaction in colorectal cancer
Lin JR, Wang S, Coy S, Chen YA, Yapp C, Tyler M, Nariya MK, Heiser CN, Lau KS, Santagata S, Sorger PK.
Cell. 2023 Jan 19. PMID: 36669472
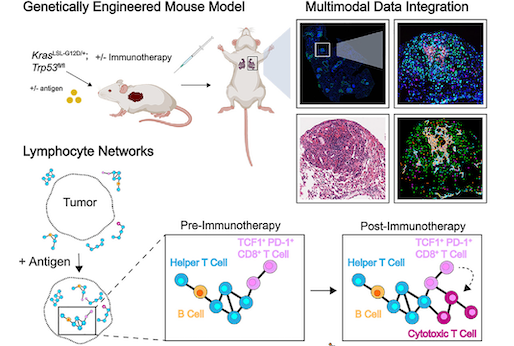
Lymphocyte networks are dynamic cellular communities in the immunoregulatory landscape of lung adenocarcinoma
Gaglia G, Burger ML, Ritch CC, Rammos D, Dai Y, Crossland GE, Tavana SZ, Warchol S, Jaeger AM, Naranjo S, Coy S, Nirmal AJ, Krueger R, Lin JR, Pfister H, Sorger PK, Jacks T, Santagata S.
Cancer Cell. 2023 Apr 8. PMID: 37059105

Evolution of delayed resistance to immunotherapy in a melanoma responder
Liu D, Lin JR, Robitschek EJ, Kasumova GG, Heyde A, Shi A, Kraya A, Zhang G, Moll T, Frederick DT, Chen YA, Wang S, Schapiro D, Ho LL, Bi K, Sahu A, Mei S, Miao B, Sharova T, Alvarez-Breckenridge C, Stocking JH, Kim T, Fadden R, Lawrence D, Hoang MP, Cahill DP, Malehmir M, Nowak MA, Brastianos PK, Lian CG, Ruppin E, Izar B, Herlyn M, Van Allen EM, Nathanson K, Flaherty KT, Sullivan RJ, Kellis M, Sorger PK, Boland GM
Nat Med. 2021 Jun. PMCID: PMC8474080

MITI minimum information guidelines for highly multiplexed tissue images
Schapiro D, Yapp C, Sokolov A, Reynolds SM, Chen YA, Sudar D, Xie Y, Muhlich J, Arias-Camison R, Arena S, Taylor AJ, Nikolov M, Tyler M, Lin JR, Burlingame EA, Human Tumor Atlas Network, Chang YH, Farhi SL, Thorsson V, Venkatamohan N, Drewes JL, Pe’er D, Gutman DA, Herrmann MD, Gehlenborg N, Bankhead P, Roland JT, Herndon JM, Snyder MP, Angelo M, Nolan G, Swedlow JR, Schultz N, Merrick DT, Mazzili SA, Cerami E, Rodig SJ, Santagata S, Sorger PK.
Nat Methods. 2022 Mar. PMCID: PMC9009186
Causal inference in medical records and complementary systems pharmacology for metformin drug repurposing towards dementia
Charpignon ML, Vakulenko-Lagun B, Zheng B, Magdamo C, Su B, Evans K, Rodriguez S, Sokolov A, Boswell S, Sheu YH, Somai M, Middleton L, Hyman BT, Betensky RA, Finkelstein SN, Welsch RE, Tzoulaki I, Blacker D, Das S, Albers MW.
Nature Communications. 2022 Dec 10. PMCID: PMC9741618

Single cell spatial analysis reveals the topology of immunomodulatory purinergic signaling in glioblastoma
Coy S, Wang S, Stopka SA, Lin JR, Yapp C, Ritch CC, Salhi L, Baker GJ, Rashid R, Baquer G, Regan M, Khadka P, Cole KA, Hwang J, Wen PY, Bandopadhayay P, Santi M, De Raedt T, Ligon KL, Agar NYR, Sorger PK, Touat M, Santagata S.
Nat Commun. 2022 Aug 16. PMCID: PMC9381513

Receptor-driven ERK pulses reconfigure MAPK signaling and enable persistence of drug-adapted BRAF-mutant melanoma cells
Gerosa L, Chidley C, Fröhlich F, Sanchez G, Lim SK, Muhlich J, Chen JY, Vallabhaneni S, Baker GJ, Schapiro D, Atanasova MI, Chylek LA, Shi T, Yi L, Nicora CD, Claas A, Ng TSC, Kohler RH, Lauffenburger DA, Weissleder R, Miller MA, Qian WJ, Wiley HS, Sorger PK.
Cell Syst. 2020 Nov 18. PMCID: PMC8009031
UnMICST: Deep learning with real augmentation for robust segmentation of highly multiplexed images of human tissues.
Yapp C, Novikov E, Jang W-D, Vallius T, Chen Y-A, Cicconet M, Maliga Z, Jacobson CA, Wei D, Santagata S, Pfister H, Sorger PK.
Communications Biology. 2022 Nov 18;5(1):1263. PMID: 36400937. PMCID: PMC9674686.
Lineage abundance estimation for SARS-CoV-2 in wastewater using transcriptome quantification techniques
Baaijens JA, Zulli A, Ott IM, Nika I, van der Lugt MJ, Petrone ME, Alpert T, Fauver JR, Kalinich CC, Vogels CBF, Breban MI, Duvallet C, McElroy KA, Ghaeli N, Imakaev M, Mckenzie-Bennett MF, Robison K, Plocik A, Schilling R, Pierson M, Littlefield R, Spencer ML, Simen BB, Yale SARS-CoV-2 Genomic Surveillance Initiative, Hanage WP, Grubaugh ND, Peccia J, Baym M.
Genome Biology. 2022 Nov 8;23(1):236. PMCID: PMC9643916
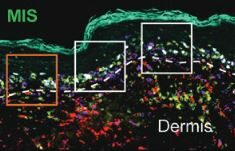
The spatial landscape of progression and immunoediting in primary melanoma at single cell resolution.
Nirmal AJ, Maliga Z, Vallius T, Quattrochi B, Chen AC, Jacobson CA, Pelletier RJ, Yapp C, Arias-Camison R, Chen Y-A, Lian CG, Murphy GF, Santagata S, Sorger PK.
Cancer Discovery. 2022 Apr 11. PMCID: PMC9167783.

Response and mechanisms of resistance to larotrectinib and selitrectinib in metastatic undifferentiated sarcoma harboring oncogenic fusion of NTRK1
Hemming ML, Nathenson MJ, Lin JR, Mei S, Du Z, Malik K, Marino-Enriquez A, Jagannathan JP, Sorger PK, Bertagnolli M, Sicinska E, Demetri GD, Santagata S.
JCO Precis Oncol. 2020 Feb 14. PMCID: PMC7055910
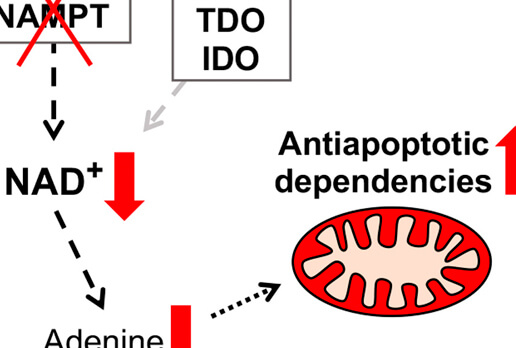
Metabolic perturbations sensitize triple-negative breast cancers to apoptosis induced by BH3 mimetics.
Daniels VW, Zoeller JJ, van Gastel N, McQueeney KE, Parvin S, Potter DS, Fell GG, Ferreira VG, Yilma B, Gupta R, Spetz J, Bhola PD, Endress JE, Harris IS, Carrilho E, Sarosiek KA, Scadden DT, Brugge JS, Letai A.
Science Signaling. 2021 Jun 8;14(686):eabc7405. doi: 10.1126/scisignal.abc7405. PMID: 34103421. PMCID: PMC8285073.

Targeting immunosuppressive macrophages overcomes PARP inhibitor resistance in BRCA1-associated triple-negative breast cancer.
Mehta, AK, Cheney, EM, Hartl, CA, Pantelidou, C, Oliwa, M, Castrillon, JA, Lin, JR, Hurst, KE, … Mittendorf EA, Sorger, PK, Shapiro, GI, Guerriero, JL.
Nature Cancer. 2021 Jan;2(1):66-82. doi: 10.1038/s43018-020-00148-7. PMID: 33738458. PMCID: PMC7963404.
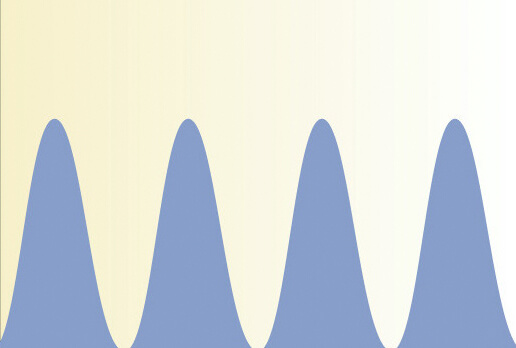
A switch in p53 dynamics marks cells that escape from DSB-induced cell cycle arrest.
Tsabar M, Mock CS, Venkatachalam V, Reyes J, Karhohs KW, Oliver TG, Regev A, Jambhekar A, Lahav G.
Cell Reports. 2020 Aug 4;32(5):107995. doi: 10.1016/j.celrep.2020.107995. 32(5):107995. PMID: 32755587; PMCID: PMC7521664.
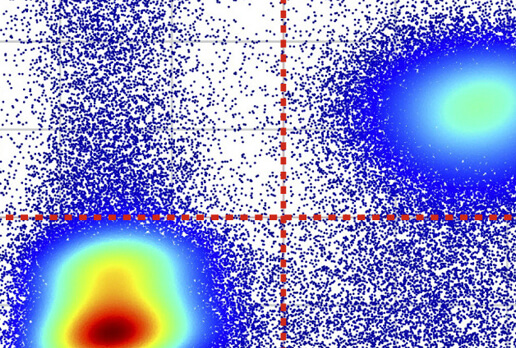
SYLARAS: A platform for the statistical analysis and visual display of systemic immunoprofiling data and its application to glioblastoma.
Baker GJ, Muhlich JL, Palaniappan SK, Moore JK, Davis SH, Santagata S, Sorger PK.
Cell Systems. 2020 Sep 23;11(3):272-285.e9. doi: 10.1016/j.cels.2020.08.001. PMID: 32898474. PMCID: PMC7565356.