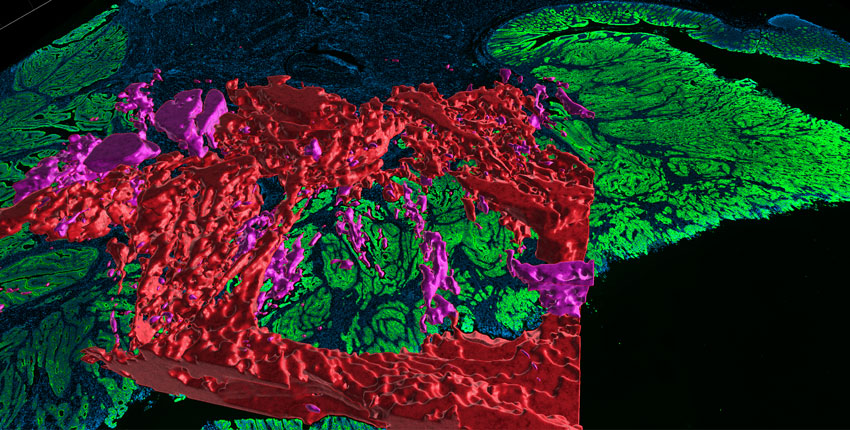
Related Publications
2576724
IXMHRBF6
colorectal cancer
1
apa-cv
50
date
desc
1
1
4133
https://labsyspharm.org/wp-content/plugins/zotpress/
%7B%22status%22%3A%22success%22%2C%22updateneeded%22%3Afalse%2C%22instance%22%3Afalse%2C%22meta%22%3A%7B%22request_last%22%3A0%2C%22request_next%22%3A0%2C%22used_cache%22%3Atrue%7D%2C%22data%22%3A%5B%7B%22key%22%3A%22U6N5QSRP%22%2C%22library%22%3A%7B%22id%22%3A2576724%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A9036456%2C%22username%22%3A%22jtefft%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fjtefft%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Loi%20et%20al.%22%2C%22parsedDate%22%3A%222024-12-02%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3ELoi%2C%20P.%2C%20Schade%2C%20A.%20E.%2C%20Rodriguez%2C%20C.%20L.%2C%20Krishnan%2C%20A.%2C%20Perurena%2C%20N.%2C%20Nguyen%2C%20V.%20T.%20M.%2C%20Xu%2C%20Y.%2C%20Watanabe%2C%20M.%2C%20Davis%2C%20R.%20A.%2C%20Gardner%2C%20A.%2C%20Pilla%2C%20N.%20F.%2C%20Mattioli%2C%20K.%2C%20Popow%2C%20O.%2C%20Gunduz%2C%20N.%2C%20Lannagan%2C%20T.%20R.%20M.%2C%20Fitzgerald%2C%20S.%2C%20Sicinska%2C%20E.%20T.%2C%20Lin%2C%20J.-R.%2C%20Tan%2C%20W.%2C%20%26%23x2026%3B%20Cichowski%2C%20K.%20%282024%29.%20Epigenetic%20and%20Oncogenic%20Inhibitors%20Cooperatively%20Drive%20Differentiation%20and%20Kill%20KRAS-Mutant%20Colorectal%20Cancers.%20%3Ci%3ECancer%20Discovery%3C%5C%2Fi%3E%2C%20%3Ci%3E14%3C%5C%2Fi%3E%2812%29%2C%202430%26%23x2013%3B2449.%20%3Ca%20class%3D%27zp-DOIURL%27%20target%3D%27_blank%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1158%5C%2F2159-8290.CD-23-0866%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1158%5C%2F2159-8290.CD-23-0866%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Epigenetic%20and%20Oncogenic%20Inhibitors%20Cooperatively%20Drive%20Differentiation%20and%20Kill%20KRAS-Mutant%20Colorectal%20Cancers%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Patrick%22%2C%22lastName%22%3A%22Loi%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Amy%20E.%22%2C%22lastName%22%3A%22Schade%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Carrie%20L.%22%2C%22lastName%22%3A%22Rodriguez%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Anjana%22%2C%22lastName%22%3A%22Krishnan%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Naiara%22%2C%22lastName%22%3A%22Perurena%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Van%20T.%20M.%22%2C%22lastName%22%3A%22Nguyen%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Yilin%22%2C%22lastName%22%3A%22Xu%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Marina%22%2C%22lastName%22%3A%22Watanabe%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Rachel%20A.%22%2C%22lastName%22%3A%22Davis%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Alycia%22%2C%22lastName%22%3A%22Gardner%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Natalie%20F.%22%2C%22lastName%22%3A%22Pilla%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Kaia%22%2C%22lastName%22%3A%22Mattioli%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Olesja%22%2C%22lastName%22%3A%22Popow%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Nuray%22%2C%22lastName%22%3A%22Gunduz%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Tamsin%20R.%20M.%22%2C%22lastName%22%3A%22Lannagan%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Samantha%22%2C%22lastName%22%3A%22Fitzgerald%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Ewa%20T.%22%2C%22lastName%22%3A%22Sicinska%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jia-Ren%22%2C%22lastName%22%3A%22Lin%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22William%22%2C%22lastName%22%3A%22Tan%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lauren%20K.%22%2C%22lastName%22%3A%22Brais%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Kevin%20M.%22%2C%22lastName%22%3A%22Haigis%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Marios%22%2C%22lastName%22%3A%22Giannakis%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Kimmie%22%2C%22lastName%22%3A%22Ng%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Sandro%22%2C%22lastName%22%3A%22Santagata%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Kristian%22%2C%22lastName%22%3A%22Helin%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Owen%20J.%22%2C%22lastName%22%3A%22Sansom%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Karen%22%2C%22lastName%22%3A%22Cichowski%22%7D%5D%2C%22abstractNote%22%3A%22Combined%20EZH2%20and%20RAS%20pathway%20inhibitors%20kill%20KRAS-mutant%20colorectal%20cancer%20cells%20and%20promote%20durable%20tumor%20regression%20in%20vivo.%20These%20agents%20function%20by%20cooperatively%20suppressing%20the%20WNT%20pathway%2C%20driving%20differentiation%2C%20and%20epigenetically%20reprogramming%20cells%20to%20permit%20the%20induction%20of%20apoptotic%20signals%2C%20which%20then%20kill%20these%20more%20differentiated%20tumor%20cells.%22%2C%22date%22%3A%222024-12-02%22%2C%22language%22%3A%22eng%22%2C%22DOI%22%3A%2210.1158%5C%2F2159-8290.CD-23-0866%22%2C%22ISSN%22%3A%222159-8290%22%2C%22url%22%3A%22%22%2C%22collections%22%3A%5B%22IXMHRBF6%22%5D%2C%22dateModified%22%3A%222024-12-04T17%3A39%3A55Z%22%7D%7D%2C%7B%22key%22%3A%229LABQV53%22%2C%22library%22%3A%7B%22id%22%3A2576724%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A5018704%2C%22username%22%3A%22AlyceChen%22%2C%22name%22%3A%22Alyce%20A%20Chen%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Falycechen%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Wala%20et%20al.%22%2C%22parsedDate%22%3A%222024-09-26%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EWala%2C%20J.%2C%20de%20Bruijn%2C%20I.%2C%20Coy%2C%20S.%2C%20Gagn%26%23xE9%3B%2C%20A.%2C%20Chan%2C%20S.%2C%20Chen%2C%20Y.-A.%2C%20Hoffer%2C%20J.%2C%20Muhlich%2C%20J.%2C%20Schultz%2C%20N.%2C%20Santagata%2C%20S.%2C%20%26amp%3B%20Sorger%2C%20P.%20K.%20%282024%29.%20%3Ci%3EIntegrating%20spatial%20profiles%20and%20cancer%20genomics%20to%20identify%20immune-infiltrated%20mismatch%20repair%20proficient%20colorectal%20cancers%3C%5C%2Fi%3E%20%5BPreprint%5D.%20bioRxiv.%20%3Ca%20class%3D%27zp-DOIURL%27%20target%3D%27_blank%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1101%5C%2F2024.09.24.614701%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1101%5C%2F2024.09.24.614701%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22preprint%22%2C%22title%22%3A%22Integrating%20spatial%20profiles%20and%20cancer%20genomics%20to%20identify%20immune-infiltrated%20mismatch%20repair%20proficient%20colorectal%20cancers%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jeremiah%22%2C%22lastName%22%3A%22Wala%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Ino%22%2C%22lastName%22%3A%22de%20Bruijn%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Shannon%22%2C%22lastName%22%3A%22Coy%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Andr%5Cu00e9anne%22%2C%22lastName%22%3A%22Gagn%5Cu00e9%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Sabrina%22%2C%22lastName%22%3A%22Chan%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Yu-An%22%2C%22lastName%22%3A%22Chen%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22John%22%2C%22lastName%22%3A%22Hoffer%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jeremy%22%2C%22lastName%22%3A%22Muhlich%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Nikolaus%22%2C%22lastName%22%3A%22Schultz%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Sandro%22%2C%22lastName%22%3A%22Santagata%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Peter%20K.%22%2C%22lastName%22%3A%22Sorger%22%7D%5D%2C%22abstractNote%22%3A%22Predicting%20the%20progression%20of%20solid%20cancers%20based%20solely%20on%20genetics%20is%20challenging%20due%20to%20the%20influence%20of%20the%20tumor%20microenvironment%20%28TME%29.%20For%20colorectal%20cancer%20%28CRC%29%2C%20tumors%20deficient%20in%20mismatch%20repair%20%28dMMR%29%20are%20more%20immune%20infiltrated%20than%20mismatch%20repair%20proficient%20%28pMMR%29%20tumors%20and%20have%20better%20prognosis%20following%20resection.%20Here%20we%20quantify%20features%20of%20the%20CRC%20TME%20by%20combining%20spatial%20profiling%20with%20genetic%20analysis%20and%20release%20our%20findings%20via%20a%20spatially%20enhanced%20version%20of%20cBioPortal%20that%20facilitates%20multi-modal%20data%20exploration%20and%20analysis.%20We%20find%20that%20%5Cu223c20%25%20of%20pMMR%20tumors%20exhibit%20similar%20levels%20of%20T%20cell%20infiltration%20as%20dMMR%20tumors%20and%20that%20this%20is%20associated%20with%20better%20survival%20but%20not%20any%20specific%20somatic%20mutation.%20These%20T%20cell-infiltrated%20pMMR%20%28tipMMR%29%20tumors%20contain%20abundant%20cells%20expressing%20PD1%20and%20PDL1%20as%20well%20as%20T%20regulatory%20cells%2C%20consistent%20with%20a%20suppressed%20immune%20response.%20Thus%2C%20like%20dMMR%20CRC%2C%20tipMMR%20CRC%20may%20benefit%20from%20immune%20checkpoint%20inhibitor%20therapy.%5CnSIGNIFICANCE%3A%20pMMR%20tumors%20with%20high%20T%20cell%20infiltration%20and%20active%20immunosuppression%20are%20identifiable%20with%20a%20mid-plex%20imaging%20assay%20whose%20clinical%20deployment%20might%20double%20the%20number%20of%20treatment-na%5Cu00efve%20CRCs%20eligible%20for%20ICIs.%20Moreover%2C%20the%20low%20tumor%20mutational%20burden%20in%20tipMMR%20CRC%20shows%20that%20MMR%20status%20is%20not%20the%20only%20factor%20promoting%20immune%20infiltration.%22%2C%22genre%22%3A%22preprint%22%2C%22repository%22%3A%22bioRxiv%22%2C%22archiveID%22%3A%22%22%2C%22date%22%3A%222024-09-26%22%2C%22DOI%22%3A%2210.1101%5C%2F2024.09.24.614701%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%22IXMHRBF6%22%5D%2C%22dateModified%22%3A%222025-04-07T22%3A47%3A35Z%22%7D%7D%2C%7B%22key%22%3A%22P742XUPD%22%2C%22library%22%3A%7B%22id%22%3A2576724%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A9036456%2C%22username%22%3A%22jtefft%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fjtefft%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Lin%20et%20al.%22%2C%22parsedDate%22%3A%222023-07%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3ELin%2C%20J.-R.%2C%20Chen%2C%20Y.-A.%2C%20Campton%2C%20D.%2C%20Cooper%2C%20J.%2C%20Coy%2C%20S.%2C%20Yapp%2C%20C.%2C%20Tefft%2C%20J.%20B.%2C%20McCarty%2C%20E.%2C%20Ligon%2C%20K.%20L.%2C%20Rodig%2C%20S.%20J.%2C%20Reese%2C%20S.%2C%20George%2C%20T.%2C%20Santagata%2C%20S.%2C%20%26amp%3B%20Sorger%2C%20P.%20K.%20%282023%29.%20High-plex%20immunofluorescence%20imaging%20and%20traditional%20histology%20of%20the%20same%20tissue%20section%20for%20discovering%20image-based%20biomarkers.%20%3Ci%3ENature%20Cancer%3C%5C%2Fi%3E%2C%20%3Ci%3E4%3C%5C%2Fi%3E%287%29%2C%201036%26%23x2013%3B1052.%20%3Ca%20class%3D%27zp-DOIURL%27%20target%3D%27_blank%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1038%5C%2Fs43018-023-00576-1%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1038%5C%2Fs43018-023-00576-1%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22High-plex%20immunofluorescence%20imaging%20and%20traditional%20histology%20of%20the%20same%20tissue%20section%20for%20discovering%20image-based%20biomarkers%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jia-Ren%22%2C%22lastName%22%3A%22Lin%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Yu-An%22%2C%22lastName%22%3A%22Chen%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Daniel%22%2C%22lastName%22%3A%22Campton%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jeremy%22%2C%22lastName%22%3A%22Cooper%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Shannon%22%2C%22lastName%22%3A%22Coy%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Clarence%22%2C%22lastName%22%3A%22Yapp%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Juliann%20B.%22%2C%22lastName%22%3A%22Tefft%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Erin%22%2C%22lastName%22%3A%22McCarty%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Keith%20L.%22%2C%22lastName%22%3A%22Ligon%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Scott%20J.%22%2C%22lastName%22%3A%22Rodig%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Steven%22%2C%22lastName%22%3A%22Reese%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Tad%22%2C%22lastName%22%3A%22George%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Sandro%22%2C%22lastName%22%3A%22Santagata%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Peter%20K.%22%2C%22lastName%22%3A%22Sorger%22%7D%5D%2C%22abstractNote%22%3A%22Precision%20medicine%20is%20critically%20dependent%20on%20better%20methods%20for%20diagnosing%20and%20staging%20disease%20and%20predicting%20drug%20response.%20Histopathology%20using%20hematoxylin%20and%20eosin%20%28H%26E%29-stained%20tissue%20%28not%20genomics%29%20remains%20the%20primary%20diagnostic%20method%20in%20cancer.%20Recently%20developed%20highly%20multiplexed%20tissue%20imaging%20methods%20promise%20to%20enhance%20research%20studies%20and%20clinical%20practice%20with%20precise%2C%20spatially%20resolved%20single-cell%20data.%20Here%2C%20we%20describe%20the%20%27Orion%27%20platform%20for%20collecting%20H%26E%20and%20high-plex%20immunofluorescence%20images%20from%20the%20same%20cells%20in%20a%20whole-slide%20format%20suitable%20for%20diagnosis.%20Using%20a%20retrospective%20cohort%20of%2074%20colorectal%20cancer%20resections%2C%20we%20show%20that%20immunofluorescence%20and%20H%26E%20images%20provide%20human%20experts%20and%20machine%20learning%20algorithms%20with%20complementary%20information%20that%20can%20be%20used%20to%20generate%20interpretable%2C%20multiplexed%20image-based%20models%20predictive%20of%20progression-free%20survival.%20Combining%20models%20of%20immune%20infiltration%20and%20tumor-intrinsic%20features%20achieves%20a%2010-%20to%2020-fold%20discrimination%20between%20rapid%20and%20slow%20%28or%20no%29%20progression%2C%20demonstrating%20the%20ability%20of%20multimodal%20tissue%20imaging%20to%20generate%20high-performance%20biomarkers.%22%2C%22date%22%3A%222023-07%22%2C%22language%22%3A%22eng%22%2C%22DOI%22%3A%2210.1038%5C%2Fs43018-023-00576-1%22%2C%22ISSN%22%3A%222662-1347%22%2C%22url%22%3A%22%22%2C%22collections%22%3A%5B%22IXMHRBF6%22%5D%2C%22dateModified%22%3A%222025-01-28T22%3A17%3A06Z%22%7D%7D%2C%7B%22key%22%3A%22YJ95V4GV%22%2C%22library%22%3A%7B%22id%22%3A2576724%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A9036456%2C%22username%22%3A%22jtefft%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fjtefft%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Lin%20et%20al.%22%2C%22parsedDate%22%3A%222023-01-19%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3ELin%2C%20J.-R.%2C%20Wang%2C%20S.%2C%20Coy%2C%20S.%2C%20Chen%2C%20Y.-A.%2C%20Yapp%2C%20C.%2C%20Tyler%2C%20M.%2C%20Nariya%2C%20M.%20K.%2C%20Heiser%2C%20C.%20N.%2C%20Lau%2C%20K.%20S.%2C%20Santagata%2C%20S.%2C%20%26amp%3B%20Sorger%2C%20P.%20K.%20%282023%29.%20Multiplexed%203D%20atlas%20of%20state%20transitions%20and%20immune%20interaction%20in%20colorectal%20cancer.%20%3Ci%3ECell%3C%5C%2Fi%3E%2C%20%3Ci%3E186%3C%5C%2Fi%3E%282%29%2C%20363-381.e19.%20%3Ca%20class%3D%27zp-DOIURL%27%20target%3D%27_blank%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.cell.2022.12.028%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.cell.2022.12.028%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Multiplexed%203D%20atlas%20of%20state%20transitions%20and%20immune%20interaction%20in%20colorectal%20cancer%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jia-Ren%22%2C%22lastName%22%3A%22Lin%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Shu%22%2C%22lastName%22%3A%22Wang%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Shannon%22%2C%22lastName%22%3A%22Coy%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Yu-An%22%2C%22lastName%22%3A%22Chen%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Clarence%22%2C%22lastName%22%3A%22Yapp%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Madison%22%2C%22lastName%22%3A%22Tyler%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Maulik%20K.%22%2C%22lastName%22%3A%22Nariya%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Cody%20N.%22%2C%22lastName%22%3A%22Heiser%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Ken%20S.%22%2C%22lastName%22%3A%22Lau%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Sandro%22%2C%22lastName%22%3A%22Santagata%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Peter%20K.%22%2C%22lastName%22%3A%22Sorger%22%7D%5D%2C%22abstractNote%22%3A%22Advanced%20solid%20cancers%20are%20complex%20assemblies%20of%20tumor%2C%20immune%2C%20and%20stromal%20cells%20characterized%20by%20high%20intratumoral%20variation.%20We%20use%20highly%20multiplexed%20tissue%20imaging%2C%203D%20reconstruction%2C%20spatial%20statistics%2C%20and%20machine%20learning%20to%20identify%20cell%20types%20and%20states%20underlying%20morphological%20features%20of%20known%20diagnostic%20and%20prognostic%20significance%20in%20colorectal%20cancer.%20Quantitation%20of%20these%20features%20in%20high-plex%20marker%20space%20reveals%20recurrent%20transitions%20from%20one%20tumor%20morphology%20to%20the%20next%2C%20some%20of%20which%20are%20coincident%20with%20long-range%20gradients%20in%20the%20expression%20of%20oncogenes%20and%20epigenetic%20regulators.%20At%20the%20tumor%20invasive%20margin%2C%20where%20tumor%2C%20normal%2C%20and%20immune%20cells%20compete%2C%20T%5Cu00a0cell%20suppression%20involves%20multiple%20cell%20types%20and%203D%20imaging%20shows%20that%20seemingly%20localized%202D%20features%20such%20as%20tertiary%20lymphoid%20structures%20are%20commonly%20interconnected%20and%20have%20graded%20molecular%20properties.%20Thus%2C%20while%20cancer%20genetics%20emphasizes%20the%20importance%20of%20discrete%20changes%20in%20tumor%20state%2C%20whole-specimen%20imaging%20reveals%20large-scale%20morphological%20and%20molecular%20gradients%20analogous%20to%20those%20in%20developing%20tissues.%22%2C%22date%22%3A%222023-01-19%22%2C%22language%22%3A%22eng%22%2C%22DOI%22%3A%2210.1016%5C%2Fj.cell.2022.12.028%22%2C%22ISSN%22%3A%221097-4172%22%2C%22url%22%3A%22%22%2C%22collections%22%3A%5B%22IXMHRBF6%22%5D%2C%22dateModified%22%3A%222024-10-25T21%3A41%3A47Z%22%7D%7D%2C%7B%22key%22%3A%22NFQRYNKP%22%2C%22library%22%3A%7B%22id%22%3A2576724%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A9036456%2C%22username%22%3A%22jtefft%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fjtefft%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Hemming%20et%20al.%22%2C%22parsedDate%22%3A%222021-03-15%22%2C%22numChildren%22%3A5%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EHemming%2C%20M.%20L.%2C%20Coy%2C%20S.%2C%20Lin%2C%20J.-R.%2C%20Andersen%2C%20J.%20L.%2C%20Przybyl%2C%20J.%2C%20Mazzola%2C%20E.%2C%20Abdelhamid%20Ahmed%2C%20A.%20H.%2C%20van%20de%20Rijn%2C%20M.%2C%20Sorger%2C%20P.%20K.%2C%20Armstrong%2C%20S.%20A.%2C%20Demetri%2C%20G.%20D.%2C%20%26amp%3B%20Santagata%2C%20S.%20%282021%29.%20HAND1%20and%20BARX1%20act%20as%20transcriptional%20and%20anatomic%20determinants%20of%20malignancy%20in%20gastrointestinal%20stromal%20tumor.%20%3Ci%3EClinical%20Cancer%20Research%3A%20An%20Official%20Journal%20of%20the%20American%20Association%20for%20Cancer%20Research%3C%5C%2Fi%3E%2C%20%3Ci%3E27%3C%5C%2Fi%3E%286%29%2C%201706%26%23x2013%3B1719.%20%3Ca%20class%3D%27zp-DOIURL%27%20target%3D%27_blank%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1158%5C%2F1078-0432.CCR-20-3538%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1158%5C%2F1078-0432.CCR-20-3538%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22HAND1%20and%20BARX1%20act%20as%20transcriptional%20and%20anatomic%20determinants%20of%20malignancy%20in%20gastrointestinal%20stromal%20tumor%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Matthew%20L.%22%2C%22lastName%22%3A%22Hemming%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Shannon%22%2C%22lastName%22%3A%22Coy%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jia-Ren%22%2C%22lastName%22%3A%22Lin%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jessica%20L.%22%2C%22lastName%22%3A%22Andersen%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Joanna%22%2C%22lastName%22%3A%22Przybyl%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Emanuele%22%2C%22lastName%22%3A%22Mazzola%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Amr%20H.%22%2C%22lastName%22%3A%22Abdelhamid%20Ahmed%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Matt%22%2C%22lastName%22%3A%22van%20de%20Rijn%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Peter%20K.%22%2C%22lastName%22%3A%22Sorger%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Scott%20A.%22%2C%22lastName%22%3A%22Armstrong%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22George%20D.%22%2C%22lastName%22%3A%22Demetri%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Sandro%22%2C%22lastName%22%3A%22Santagata%22%7D%5D%2C%22abstractNote%22%3A%22PURPOSE%3A%20Gastrointestinal%20stromal%20tumor%20%28GIST%29%20arises%20from%20interstitial%20cells%20of%20Cajal%20%28ICC%29%20or%20their%20precursors%2C%20which%20are%20present%20throughout%20the%20gastrointestinal%20tract.%20Although%20gastric%20GIST%20is%20commonly%20indolent%20and%20small%20intestine%20GIST%20more%20aggressive%2C%20a%20molecular%20understanding%20of%20disease%20behavior%20would%20inform%20therapy%20decisions%20in%20GIST.%20Although%20a%20core%20transcription%20factor%20%28TF%29%20network%20is%20conserved%20across%20GIST%2C%20accessory%20TFs%20HAND1%20and%20BARX1%20are%20expressed%20in%20a%20disease%20state-specific%20pattern.%20Here%2C%20we%20characterize%20two%20divergent%20transcriptional%20programs%20maintained%20by%20HAND1%20and%20BARX1%2C%20and%20evaluate%20their%20association%20with%20clinical%20outcomes.%5CnEXPERIMENTAL%20DESIGN%3A%20We%20evaluated%20RNA%20sequencing%20and%20TF%20chromatin%20immunoprecipitation%20with%20sequencing%20in%20GIST%20samples%20and%20cultured%20cells%20for%20transcriptional%20programs%20associated%20with%20HAND1%20and%20BARX1.%20Multiplexed%20tissue-based%20cyclic%20immunofluorescence%20and%20IHC%20evaluated%20tissue-%20and%20cell-level%20expression%20of%20TFs%20and%20their%20association%20with%20clinical%20factors.%5CnRESULTS%3A%20We%20show%20that%20HAND1%20is%20expressed%20in%20aggressive%20GIST%2C%20modulating%20KIT%20and%20core%20TF%20expression%20and%20supporting%20proliferative%20cellular%20programs.%20In%20contrast%2C%20BARX1%20is%20expressed%20in%20indolent%20and%20micro-GISTs.%20HAND1%20and%20BARX1%20expression%20were%20superior%20predictors%20of%20relapse-free%20survival%2C%20as%20compared%20with%20standard%20risk%20stratification%2C%20and%20they%20predict%20progression-free%20survival%20on%20imatinib.%20Reflecting%20the%20developmental%20origins%20of%20accessory%20TF%20programs%2C%20HAND1%20was%20expressed%20solely%20in%20small%20intestine%20ICCs%2C%20whereas%20BARX1%20expression%20was%20restricted%20to%20gastric%20ICCs.%5CnCONCLUSIONS%3A%20Our%20results%20define%20anatomic%20and%20transcriptional%20determinants%20of%20GIST%20and%20molecular%20origins%20of%20clinical%20phenotypes.%20Assessment%20of%20HAND1%20and%20BARX1%20expression%20in%20GIST%20may%20provide%20prognostic%20information%20and%20improve%20clinical%20decisions%20on%20the%20administration%20of%20adjuvant%20therapy.%22%2C%22date%22%3A%222021-03-15%22%2C%22language%22%3A%22eng%22%2C%22DOI%22%3A%2210.1158%5C%2F1078-0432.CCR-20-3538%22%2C%22ISSN%22%3A%221557-3265%22%2C%22url%22%3A%22%22%2C%22collections%22%3A%5B%22IXMHRBF6%22%5D%2C%22dateModified%22%3A%222025-01-29T10%3A34%3A24Z%22%7D%7D%2C%7B%22key%22%3A%22P7LYERJF%22%2C%22library%22%3A%7B%22id%22%3A2576724%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A9036456%2C%22username%22%3A%22jtefft%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fjtefft%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Bhola%20et%20al.%22%2C%22parsedDate%22%3A%222020-06-16%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EBhola%2C%20P.%20D.%2C%20Ahmed%2C%20E.%2C%20Guerriero%2C%20J.%20L.%2C%20Sicinska%2C%20E.%2C%20Su%2C%20E.%2C%20Lavrova%2C%20E.%2C%20Ni%2C%20J.%2C%20Chipashvili%2C%20O.%2C%20Hagan%2C%20T.%2C%20Pioso%2C%20M.%20S.%2C%20McQueeney%2C%20K.%2C%20Ng%2C%20K.%2C%20Aguirre%2C%20A.%20J.%2C%20Cleary%2C%20J.%20M.%2C%20Cocozziello%2C%20D.%2C%20Sotayo%2C%20A.%2C%20Ryan%2C%20J.%2C%20Zhao%2C%20J.%20J.%2C%20%26amp%3B%20Letai%2C%20A.%20%282020%29.%20High-throughput%20dynamic%20BH3%20profiling%20may%20quickly%20and%20accurately%20predict%20effective%20therapies%20in%20solid%20tumors.%20%3Ci%3EScience%20Signaling%3C%5C%2Fi%3E%2C%20%3Ci%3E13%3C%5C%2Fi%3E%28636%29.%20%3Ca%20class%3D%27zp-DOIURL%27%20target%3D%27_blank%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1126%5C%2Fscisignal.aay1451%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1126%5C%2Fscisignal.aay1451%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22High-throughput%20dynamic%20BH3%20profiling%20may%20quickly%20and%20accurately%20predict%20effective%20therapies%20in%20solid%20tumors%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Patrick%20D.%22%2C%22lastName%22%3A%22Bhola%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Eman%22%2C%22lastName%22%3A%22Ahmed%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jennifer%20L.%22%2C%22lastName%22%3A%22Guerriero%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Ewa%22%2C%22lastName%22%3A%22Sicinska%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Emily%22%2C%22lastName%22%3A%22Su%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Elizaveta%22%2C%22lastName%22%3A%22Lavrova%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jing%22%2C%22lastName%22%3A%22Ni%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Otari%22%2C%22lastName%22%3A%22Chipashvili%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Timothy%22%2C%22lastName%22%3A%22Hagan%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Marissa%20S.%22%2C%22lastName%22%3A%22Pioso%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Kelley%22%2C%22lastName%22%3A%22McQueeney%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Kimmie%22%2C%22lastName%22%3A%22Ng%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Andrew%20J.%22%2C%22lastName%22%3A%22Aguirre%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22James%20M.%22%2C%22lastName%22%3A%22Cleary%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22David%22%2C%22lastName%22%3A%22Cocozziello%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Alaba%22%2C%22lastName%22%3A%22Sotayo%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jeremy%22%2C%22lastName%22%3A%22Ryan%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jean%20J.%22%2C%22lastName%22%3A%22Zhao%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Anthony%22%2C%22lastName%22%3A%22Letai%22%7D%5D%2C%22abstractNote%22%3A%22Despite%20decades%20of%20effort%2C%20the%20sensitivity%20of%20patient%20tumors%20to%20individual%20drugs%20is%20often%20not%20predictable%20on%20the%20basis%20of%20molecular%20markers%20alone.%20Therefore%2C%20unbiased%2C%20high-throughput%20approaches%20to%20match%20patient%20tumors%20to%20effective%20drugs%2C%20without%20requiring%20a%20priori%20molecular%20hypotheses%2C%20are%20critically%20needed.%20Here%2C%20we%20improved%20upon%20a%20method%20that%20we%20previously%20reported%20and%20developed%20called%20high-throughput%20dynamic%20BH3%20profiling%20%28HT-DBP%29.%20HT-DBP%20is%20a%20microscopy-based%2C%20single-cell%20resolution%20assay%20that%20enables%20chemical%20screens%20of%20hundreds%20to%20thousands%20of%20candidate%20drugs%20on%20freshly%20isolated%20tumor%20cells.%20The%20method%20identifies%20chemical%20inducers%20of%20mitochondrial%20apoptotic%20signaling%2C%20a%20mechanism%20of%20cell%20death.%20HT-DBP%20requires%20only%2024%20hours%20of%20ex%20vivo%20culture%2C%20which%20enables%20a%20more%20immediate%20study%20of%20fresh%20primary%20tumor%20cells%20and%20minimizes%20adaptive%20changes%20that%20occur%20with%20prolonged%20ex%20vivo%20culture.%20Effective%20compounds%20identified%20by%20HT-DBP%20induced%20tumor%20regression%20in%20genetically%20engineered%20and%20patient-derived%20xenograft%20%28PDX%29%20models%20of%20breast%20cancer.%20We%20additionally%20found%20that%20chemical%20vulnerabilities%20changed%20as%20cancer%20cells%20expanded%20ex%20vivo.%20Furthermore%2C%20using%20PDX%20models%20of%20colon%20cancer%20and%20resected%20tumors%20from%20colon%20cancer%20patients%2C%20our%20data%20demonstrated%20that%20HT-DBP%20could%20be%20used%20to%20generate%20personalized%20pharmacotypes.%20Thus%2C%20HT-DBP%20appears%20to%20be%20an%20ex%20vivo%20functional%20method%20with%20sufficient%20scale%20to%20simultaneously%20function%20as%20a%20companion%20diagnostic%2C%20therapeutic%20personalization%2C%20and%20discovery%20tool.%22%2C%22date%22%3A%222020-06-16%22%2C%22language%22%3A%22eng%22%2C%22DOI%22%3A%2210.1126%5C%2Fscisignal.aay1451%22%2C%22ISSN%22%3A%221937-9145%22%2C%22url%22%3A%22%22%2C%22collections%22%3A%5B%22IXMHRBF6%22%5D%2C%22dateModified%22%3A%222025-04-22T15%3A05%3A24Z%22%7D%7D%5D%7D
Loi, P., Schade, A. E., Rodriguez, C. L., Krishnan, A., Perurena, N., Nguyen, V. T. M., Xu, Y., Watanabe, M., Davis, R. A., Gardner, A., Pilla, N. F., Mattioli, K., Popow, O., Gunduz, N., Lannagan, T. R. M., Fitzgerald, S., Sicinska, E. T., Lin, J.-R., Tan, W., … Cichowski, K. (2024). Epigenetic and Oncogenic Inhibitors Cooperatively Drive Differentiation and Kill KRAS-Mutant Colorectal Cancers.
Cancer Discovery,
14(12), 2430–2449.
https://doi.org/10.1158/2159-8290.CD-23-0866
Wala, J., de Bruijn, I., Coy, S., Gagné, A., Chan, S., Chen, Y.-A., Hoffer, J., Muhlich, J., Schultz, N., Santagata, S., & Sorger, P. K. (2024).
Integrating spatial profiles and cancer genomics to identify immune-infiltrated mismatch repair proficient colorectal cancers [Preprint]. bioRxiv.
https://doi.org/10.1101/2024.09.24.614701
Lin, J.-R., Chen, Y.-A., Campton, D., Cooper, J., Coy, S., Yapp, C., Tefft, J. B., McCarty, E., Ligon, K. L., Rodig, S. J., Reese, S., George, T., Santagata, S., & Sorger, P. K. (2023). High-plex immunofluorescence imaging and traditional histology of the same tissue section for discovering image-based biomarkers.
Nature Cancer,
4(7), 1036–1052.
https://doi.org/10.1038/s43018-023-00576-1
Lin, J.-R., Wang, S., Coy, S., Chen, Y.-A., Yapp, C., Tyler, M., Nariya, M. K., Heiser, C. N., Lau, K. S., Santagata, S., & Sorger, P. K. (2023). Multiplexed 3D atlas of state transitions and immune interaction in colorectal cancer.
Cell,
186(2), 363-381.e19.
https://doi.org/10.1016/j.cell.2022.12.028
Hemming, M. L., Coy, S., Lin, J.-R., Andersen, J. L., Przybyl, J., Mazzola, E., Abdelhamid Ahmed, A. H., van de Rijn, M., Sorger, P. K., Armstrong, S. A., Demetri, G. D., & Santagata, S. (2021). HAND1 and BARX1 act as transcriptional and anatomic determinants of malignancy in gastrointestinal stromal tumor.
Clinical Cancer Research: An Official Journal of the American Association for Cancer Research,
27(6), 1706–1719.
https://doi.org/10.1158/1078-0432.CCR-20-3538
Bhola, P. D., Ahmed, E., Guerriero, J. L., Sicinska, E., Su, E., Lavrova, E., Ni, J., Chipashvili, O., Hagan, T., Pioso, M. S., McQueeney, K., Ng, K., Aguirre, A. J., Cleary, J. M., Cocozziello, D., Sotayo, A., Ryan, J., Zhao, J. J., & Letai, A. (2020). High-throughput dynamic BH3 profiling may quickly and accurately predict effective therapies in solid tumors.
Science Signaling,
13(636).
https://doi.org/10.1126/scisignal.aay1451