Identifying the determinants of sensitivity and resistance to small molecule kinase inhibitors
Kinase inhibitors are an intensively studied class of therapeutics with many remaining unknowns – LSP investigators use them to develop new approaches for identifying targets and to study the factors determining drug resistance and sensitivity.
Inhibitors of the roughly 540 human protein and lipid kinases are the most intensively developed and studied class of therapeutic drugs. Kinase inhibitors such as imatinib (for treatment of chronic myeloid leukemia), vemurafenib (for treatment of BRAF-mutant melanoma), and erlotinib (for treatment of EGFR-mutant lung cancer) were the first breakthrough “targeted” drugs invented for genetically defined cancers. Kinase inhibitors are also important in the treatment of inflammatory diseases. However, a primary limitation in the use of these drugs is the frequent development of resistance, which involves both a short-term adaptive resistance and genetically determined long-term resistance. Resistance mechanisms involve many of the homeostatic processes that allow cells to survive and proliferate in the presence of internal and external stress and are, therefore, interesting in their own right. By studying precisely how drug resistance arises in cell lines, animal models, and humans, we hope to prevent its emergence using newly developed drugs or combination therapies. This project is ideal for cancer biologists and oncologists interested in fundamentally improving our approach to small molecule therapy and increasing the durability of therapeutic responses. Clinical trials are underway making use of some of the insights arising from this project.
Associated with:
The HMS LINCS Center collects and disseminates data and analytical tools needed to understand how human cells respond to perturbation by drugs, the environment, and mutation. Part of the NIH Library of Integrated Network-based Cellular Signatures (LINCS) Program.
Selected Publications:
Ferguson FM, Doctor ZM, Ficarro SB, Browne CM, Marto JA, Johnson JL, Yaron TM, Cantley LC, Kim ND, Sim T, Berberich MJ, Kalocsay M, Sorger PK, Gray NS. Discovery of Covalent CDK14 Inhibitors with Pan-TAIRE Family Specificity. Cell Chem Biol. 2019 Jun 20;26(6):804-817.e12. PMCID: PMC6588450.
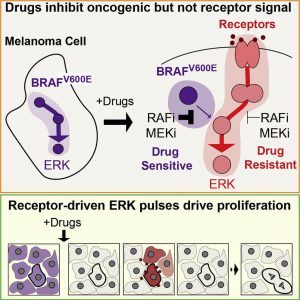
Gerosa L, Chidley C, Fröhlich F, Sanchez G, Lim SK, Muhlich J, Chen J-Y, Vallabhaneni S, Baker GJ, Schapiro D, Atanasova MI, Chylek LA, Shi T, Yi L, Nicora CD, Claas A, Ng TSC, Kohler RH, Lauffenburger DA, Weissleder R, Miller MA, Qian W-J, Wiley HS, Sorger PK. Receptor-Driven ERK Pulses Reconfigure MAPK Signaling and Enable Persistence of Drug-Adapted BRAF-Mutant Melanoma Cells. Cell Syst. 2020 Nov 18;11(5):478-494.e9. PMCID: PMC8009031.
Hafner M, Niepel M, Chung M, Sorger PK. Growth rate inhibition metrics correct for confounders in measuring sensitivity to cancer drugs. Nat Methods. 2016 Jun 1;13(6):521–527. PMCID: PMC4887336.
Hafner M, Mills CE, Subramanian K, Chen C, Chung M, Boswell SA, Everley RA, Liu C, Walmsley CS, Juric D, Sorger PK. Multiomics Profiling Establishes the Polypharmacology of FDA-Approved CDK4/6 Inhibitors and the Potential for Differential Clinical Activity. Cell Chem Biol. 2019 Aug 15;26(8):1067-1080.e8. PMCID: PMC6936329.
Fröhlich F, Gerosa L, Muhlich J, Sorger PK. Mechanistic model of MAPK signaling reveals how allostery and rewiring contribute to drug resistance. Mol Syst Biol. 2023 Feb 10;19(2):e10988. PMCID: PMC9912026