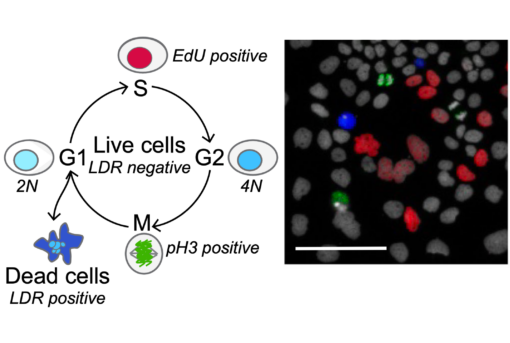
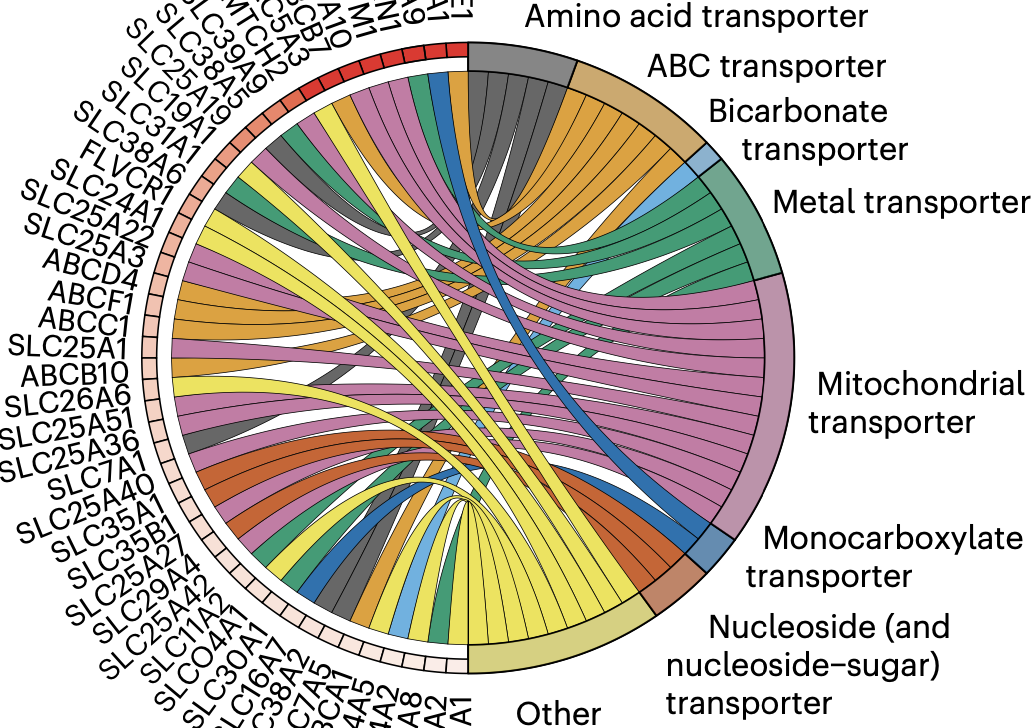
Systems Pharmacology

By pairing computational and experimental approaches, LSP investigators enable a rational, systems-level approach for evaluating preclinical compounds.
Despite the high costs associated with drug development, only 10% of preclinical compounds pass clinical trials. This drop-off is partially because most preclinical studies reduce a disease to its simplest components, focusing on a few specific targets in a limited number of disease models. Although this reductionist approach provides useful results, it overlooks the fact that within an organism, most drugs interact with multiple targets at the same time— a phenomenon known as polypharmacology.
To address this gap, LSP researchers use quantitative systems pharmacology, combining traditional drug development approaches with engineering and computation to model how drugs will behave in the body.
Since 2014, LSP investigators have developed numerous systems pharmacology methods for evaluating preclinical compounds. These efforts help clarify how polypharmacology influences drug action, uncover sources of drug sensitivity and resistance, and inform clinical trial design. Ultimately, these efforts aim to provide a quantitative basis for identifying preclinical compounds with the greatest potential for clinical success.